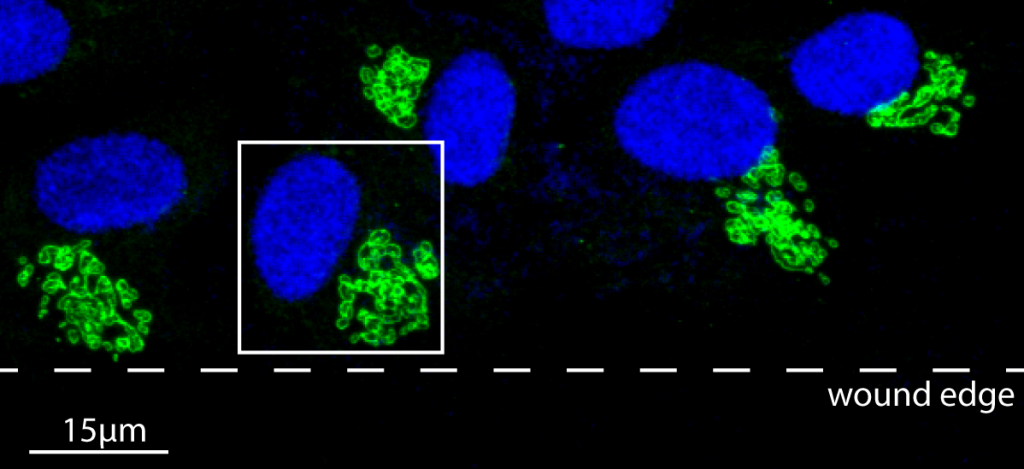
We developed image analysis software to accurately track irregularly shaped objects (such as Golgi bodies or other cell organelles), and we connect this with nuclei tracking software to quantify the behavior of the nuclei-Golgi body axis as an intracellular marker of cell polarization. We find that this marker of cell polarization displays much more sensitivity to substrate anisotropy than the nuclei shape, suggesting that models for cell polarization should be updated to include these effects. We also have developed a software package to determine mechanisms for superdiffusive cell motion on 2D substrates, through a careful analysis of cell trajectories compared to a generalized self-propelled model.
Manning group participants: Giuseppe Passucci, M Manning.
Collaborators: M. Brasch, J. Henderson (SU BMCE and SBI), Anushree Gulvady and Chris Turner, SUNY Upstate Medical University. Funding: NSF- BMMB